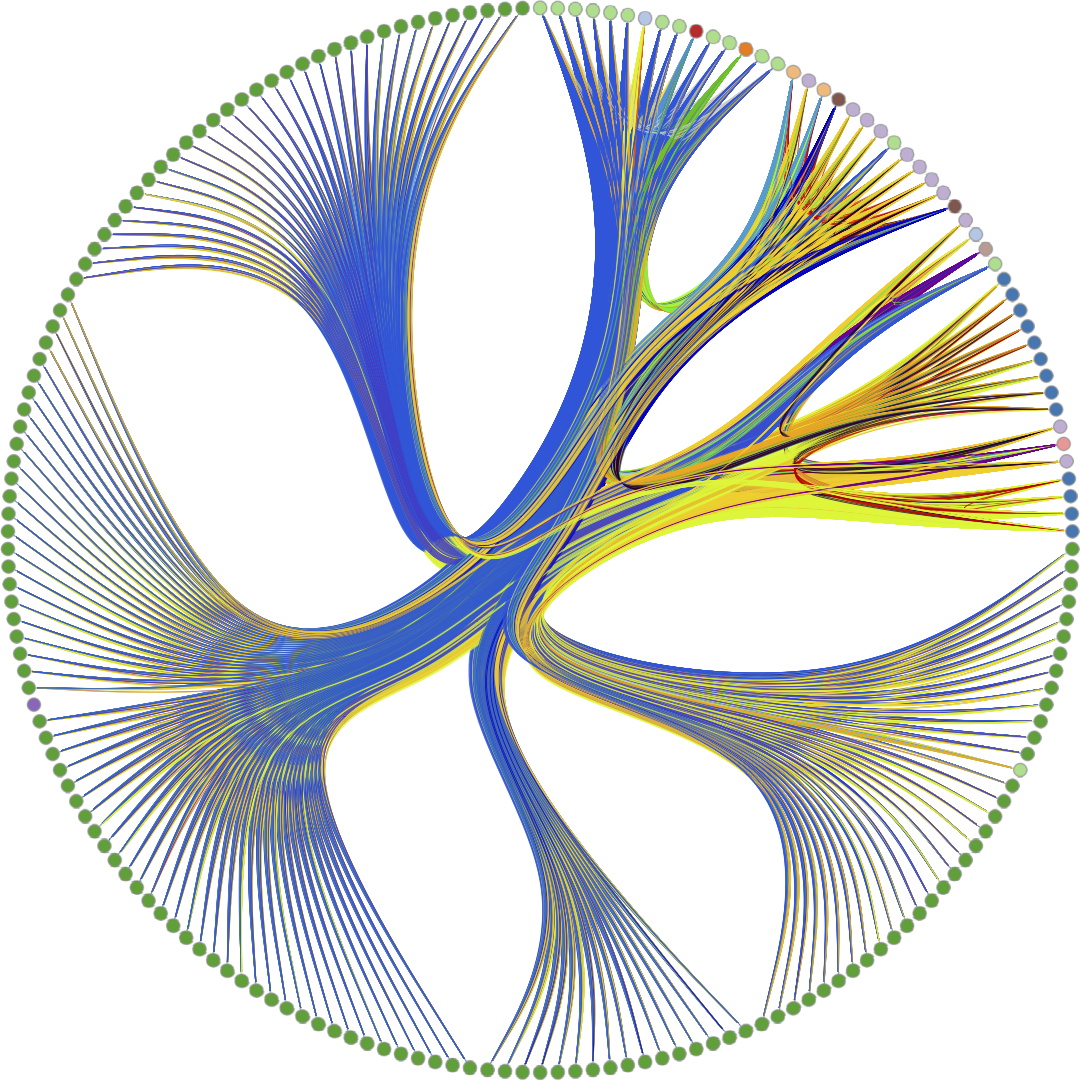
microbial recombination
how does recombination shape microbial genome evolution?
My PhD work on building a statistical physics framework for understanding how dietary polymers influence gut function sparked my interest in quantifying the evolution of the gut’s microbial inhabitants. Therefore, my postdoctoral work with Edo Kussell at the Center for Genomics and Systems Biology at New York University has focused on microbial population genetics and bioinformatics. As a Simons Postdoctoral Fellow of the LSRF at NYU, I am studying the myriad ways by which homologous recombination impacts microbial evolution. While microbial evolution was once thought to be solely the consequence of mutation, it has become increasingly apparent that recombination plays a key role in reshaping the microbial genome. It can dramatically alter the evolutionary trajectories of microbial populations, as it is involved in the proliferation of antibiotic resistance, antigen variation, and adaptation to the host niche. Despite its importance, quantifying homologous recombination rates in bacteria and viruses has remained challenging due to its obfuscation of clonal relationships, which stymies classical phylogenetic methods. With the Kussell Lab, I have been working on extending and applying their recently developed method for inferring homologous recombination rates in bacteria (mcorr), which relies on the measurement of statistical correlation functions in large-scale sequencing data, to a range of problems in microbial evolution. Specifically, in recent work we have quantified the variation in homologous recombination across core and accessory genes (essential genes present in all strains of a given microbial species and niche-adaptive genes present in a subset of strains). We analyzed >100,000 genomes from 12 bacterial species and found that, despite being the most conserved part of the genome, core genes often have higher homologous recombination rates than accessory genes. Quantifying this intra-genomic variation will allow us to better understand how selective pressures interact with recombination to shape different classes of genes.
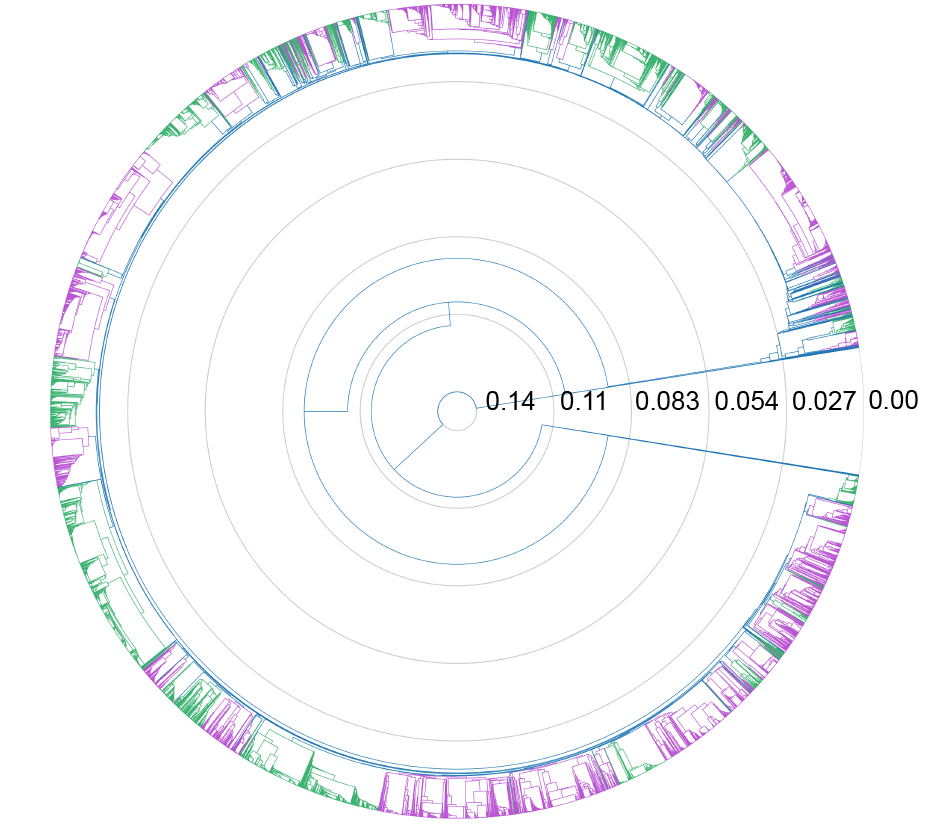
In a separate project, I have adapted the theory and computational approach of our method to measure recombination rates in RNA viruses and, particularly, SARS-like coronaviruses (paper here and analysis pipeline on GitHub). This method allows for the rapid analysis of hundreds of thousands of whole genome sequences. Using this technique, we have uncovered new insights into the clonal relationships of SARS-like coronavirus sequences and the recombination dynamics of SARS-like coronavirus gene pools. This new tool will allow us to better understand and predict both SARS-like coronavirus and RNA virus evolution.